

The planting of transgenic rice has aroused ongoing controversy, due to the public anxiety surrounding the potential risk of transgenic rice to health and the environment. The soil microbial community plays an important environmental role in the plant-soil-microbe system; however, few studies have focused on the effect of transgenic rice on the soil rhizospheric microbiome. We labeled transgenic gene rice (TT51, transformed with Cry1Ab/1Ac gene), able to produce the Bt (Bacillus thuringiensis) toxin, its parental variety (Minghui 63), and a non-parental variety (9931) with 13CO2. The DNA of the associated soil rhizospheric microbes was extracted, subjected to density gradient centrifugation, followed by high-throughput sequencing of bacterial 16S rRNA gene. Unweighted unifrac analysis of the sequencing showed that transgenic rice did not significantly change the soil bacterial community structure compared with its parental variety. The order Opitutales, affiliated to phylum Verrucomicrobia and order Sphingobacteriales, was the main group of labeled bacteria in soil planted with the transgenic and parental varieties, while the orders Pedosphaerales, Chthoniobacteraceae, also affiliated to Verrucomicrobia, and the genus Geobacter, affiliated to class Deltaproteobacteria, dominated in the soil of the non-parental rice variety. The non-significant difference in soil bacterial community structure of labeled microbes between the transgenic and parental varieties, but the comparatively large difference with the non-parental variety, suggests a limited effect of planting transgenic Bt rice on the soil microbiome.
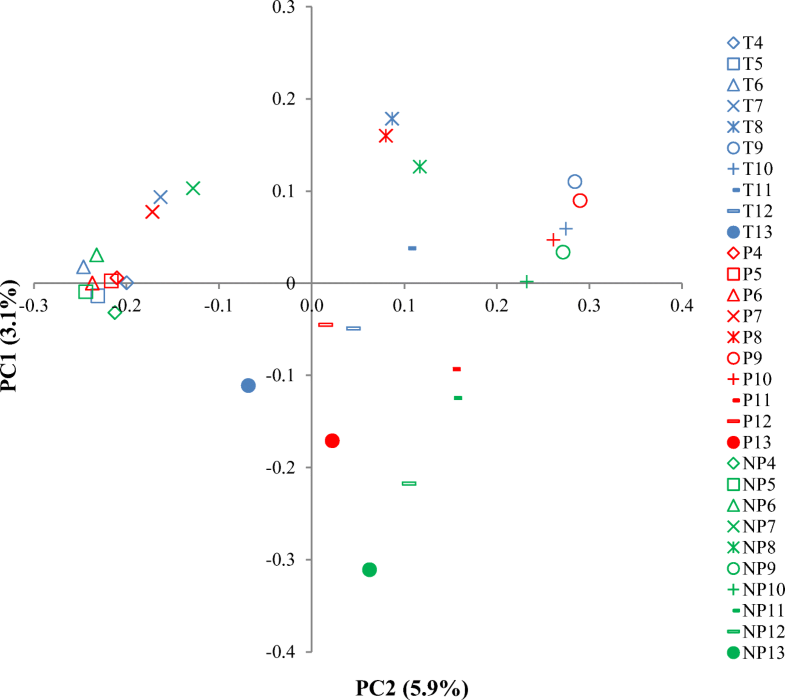
The unweighted unifrac analysis of high-throughput sequencing for each treatment and each fraction for the 13CO2-labeled treatments. The fractions after ultracentrifugation from high to low buoyant densities (4–13). The values are the average of triplicate analyses